Abstract
Introduction:
Infant acute lymphoblastic leukemia (ALL) is an aggressive subtype of leukemia with low rates of survival. Rearrangements involving the KMT2A gene are associated with poor prognosis for infants with this cancer. Although many patients with KMT2A rearrangements (KMT2A-r) ultimately relapse, some do not. The molecular basis for this distinction has not yet been determined. There is evidence to suggest that cancers with distinct KMT2A fusion partners show distinct patterns of gene expression, though these differences have not previously been linked to prognostic outcomes. Here we utilize single-cell genomic analysis for infant ALL samples with and without KMT2A-r taken at diagnosis to explore transcriptional dynamics and identify gene expression programs with potential prognostic value.
Methods:
We performed 10x Chromium single-cell multiome sequencing to obtain both gene expression (scRNA-seq) and chromatin accessibility (scATAC-seq) data for blood and/or bone marrow samples obtained from a total of 34 infant ALL patients at time of diagnosis. Of these, 19 KMT2A-r patients later relapsed, 6 KMT2A-r patients did not relapse, and 9 patients did not show KMT2A rearrangement. Sequencing data were processed and aggregated with cellranger-arc, with normalization and differential gene expression performed using the Seurat package for gene expression analysis. Differential gene expression was performed between the three groups described above, (KMT2A-r + relapse, KMT2A-r no relapse, no KMT2A-r), then further subdivided for KMT2A-r cases based on the KMT2A partner gene (MLLT1 n=13 or AFF1 n=12).
Results:
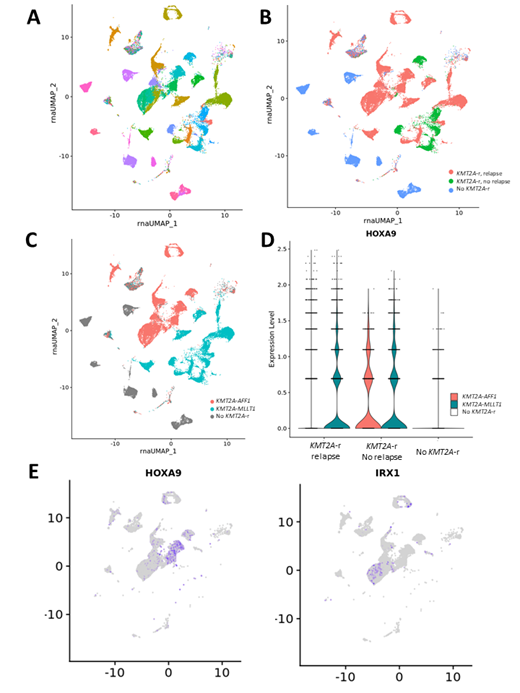
In preliminary scRNA- and scATAC-seq in infant ALL patients, we had observed that cancer cells from individual patients tend to cluster separately, while non-blast cell populations showed similar interindividual patterns, suggesting a high degree of divergence among blast cell transcriptional programs. We replicated this finding here in larger patient samples, demonstrating no overt similarity based on KMT2A-r status or whether patients later relapsed. Strikingly, samples did broadly cluster according to KMT2A rearrangement partner gene, indicating that this appears to be a major driver of the transcriptional program in infant ALL patients. Differential expression analysis within the KMT2A-MLLT1 and KMT2A-AFF1 samples separately revealed expression of other genes associated with particular KMT2A fusion genes. Specifically, we observed that expression of HOXA genes was mostly restricted to KMT2A-MLLT1 samples. Consistent with previous results, we observed that within KMT2A-AFF1 samples there were two distinct groups of cells with mutually exclusive expression of HOXA9 or IRX1. Furthermore, the limited HOX gene expression observed in KMT2A-AFF1 samples was enriched in patients who did not relapse, supporting previous observations that the KMT2A-AFF1 samples expressing IRX1 comprise a more aggressive leukemia.
Conclusions:
Utilizing a single-cell transcriptomic approach enabled us to identify gene expression programs associated with good and poor prognosis in KMT2A-r infant ALL cases. We found that the KMT2A fusion partner gene appears to drive a large portion of the transcriptional heterogeneity observed across KMT2A-r samples. By treating the KMT2A-MLLT1 and KMT2A-AFF1 samples separately and exploring transcriptional dynamics within these groups, we identified transcriptional differences between patients who did and did not relapse in the presence of a KMT2A-AFF1 fusion. We are continuing to explore these data to identify prognostic markers in KMT2A-MLLT1 patients.
Figure 1. Mutually exclusive gene expression programs distinguish subsets of infant ALL samples. A) Uniform manifold approximation and projection (UMAP) visualization of gene expression in individual cells across all patient samples at diagnosis, each color corresponding to a single patient (n=34). B) Cells colored according to KMT2A-r status and relapse or lack thereof. C) Cells colored according to KMT2A fusion partner gene. D) HOXA9 expression among KMT2A-AFF1 patients is strongly biased to patients that do not relapse. E) In KMT2A-AFF1 samples, HOXA9 and IRX1 show mutually exclusive expression patterns.
Brown: Novartis: Membership on an entity's Board of Directors or advisory committees; Takeda: Membership on an entity's Board of Directors or advisory committees; Amgen: Membership on an entity's Board of Directors or advisory committees; Kura: Membership on an entity's Board of Directors or advisory committees; KIte: Membership on an entity's Board of Directors or advisory committees.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal